Enhancer Journal Club: Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes
I've been thinking a lot about super-enhancers recently and I found this paper1 from Jennifer Mitchell's lab very interesting, especially as whether and how super-enhancers are distinct from "normal" enhancers has been generating a lot of discussion in the enhancer/gene-regulation literature recently2,3
Here's the abstract:
Transcriptional enhancers are critical for maintaining cell-type–specific gene expression and driving cell fate changes during development. Highly transcribed genes are often associated with a cluster of individual enhancers such as those found in locus control regions. Recently, these have been termed stretch enhancers or super-enhancers, which have been predicted to regulate critical cell identity genes.We employed a CRISPR/Cas9-mediated deletion approach to study the func- tion of several enhancer clusters (ECs) and isolated enhancers in mouse embryonic stem (ES) cells. Our results reveal that the effect of deleting ECs, also classified as ES cell super-enhancers, is highly variable, resulting in target gene expression reductions ranging from 12% to as much as 92%. Partial deletions of these ECs which removed only one enhancer or a subcluster of enhancers revealed partially redundant control of the regulated gene by multiple enhancers within the larg- er cluster. Many highly transcribed genes in ES cells are not associated with a super-enhancer; furthermore, super-enhanc- er predictions ignore 81% of the potentially active regulatory elements predicted by cobinding of five or more pluripotency-associated transcription factors. Deletion of these additional enhancer regions revealed their robust regula- tory role in gene transcription. In addition, select super-enhancers and enhancers were identified that regulated clusters of paralogous genes. We conclude that, whereas robust transcriptional output can be achieved by an isolated enhancer, clusters of enhancers acting on a common target gene act in a partially redundant manner to fine tune transcriptional output.
Super-enhancers are clusters of enhancers that are highly enriched for certain specific transcription factors, transcriptional regulators or chromatin marks. They have been associated with the regulation of important developmental genes, and one key question surrounding them is whether they represent something functionally distinct from the regular, "common-or-garden" enhancer. In this paper Moorthy et al. set out to address this question by using genome-editing techniques to delete some super-enhancers and examine the effect on the expression of nearby genes. They do this using a very clever experimental approach first applied by Bing Ren's lab4. Essentially, since a true enhancer should only regulate genes lying on the same chromosome, you delete either the maternal or the paternal copy of the gene from a hybrid mouse. You then measure gene expression and look for genes where only the maternal copy is supressed (if the maternal enhancer was deleted) or only the paternal copy is supressed (if the paternal enhancer was deleted).
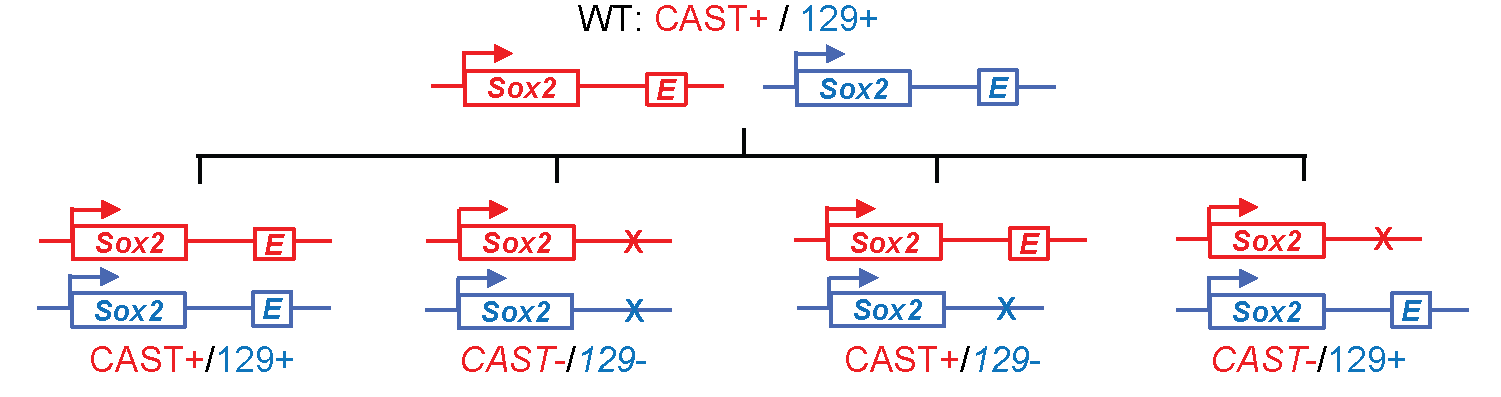
Fig: By knocking out an enhancer from either the maternal or paternal genome in a hybrid mouse, it is possible to isolate the immediate gene targets of the enhancer (on the same chromsosome) and exclude secondary effects. Figure reproduced with modifications from Li, Y. et al., CRISPR Reveals a Distal Super-Enhancer Required for Sox2 Expression in Mouse Embryonic Stem Cells. PLOS One 9:e114485 (2014).
I really enjoyed the Li et al. paper, so I was excited to see that another lab had taken this approach and applied it to a much larger variety of enhancers and super-enhancers. The results are pretty interesting - just like in the Li et al. paper, the majority (8/11) of tested enhancer regions turn out to regulate just a single gene, and usually one that lies fairly close on the chromosome. This is actually quite intriguing in the light of recent reports that at least some enhancers can act from essentially any position within their topological domain. Since the super-enhancers investigated generally share their topological domains with multiple other genes, it's possible that some factor other than spatial proximity prevents the super-enhancers from acting on more than one or two targets.
Another nice approach this paper takes is to study the effect of deleting individual enhancers from two different clusters of enhancers. In both cases, they observe partial redundancy, such that removing a few individual enhancers from a cluster has much less dramatic effects on the expression of a target gene than removing the entire cluster. This is more-or-less in line with the recent results from the alpha-globlin locus5, insofar as that both studies suggest super-enhancers are not formed by co-operation between their constitutive enhancer elements, although at alpha-globin the individual enhancers have addititve effects rather than partially redundant ones.
So finally back to the bottom line, are super-enhancers different from regular enhancers? The authors of this paper make the point that enhancer clusters not identified as super-enhancers can have very large effects on target gene expression when they are deleted, showing that super-enhancer predictions can and will exclude regulatory regions with strong effects. Given that algorithms for detecting super-enhancers generally identify just a few hundred such elements per cell type, I'm not sure it is suprising that these predictions miss out some important elements. For me the important point is that enhancer predictions often generate large numbers of false negatives - regions that turn out not to affect the expression of nearby genes when they are deleted. I think the jury is still out on whether super-enhancer predictions better or worse at excluding these kinds of false negative enhancers. This paper does not examine enough of either class of regulatory element to make strong claims about which ways of identifying enhancers are better, but it certainly shows that we have much more to learn, and it's a great showcase for the power of heterozygous regulatory element deletions in hybrid mice.
- (Moorthy S. et al., Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Research 27:246–258 (2016).) ↩
- (Shin H. et al., Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nature Genetics 48:904–911 (2016).) ↩
- (Pott & Lieb, What are super-enhancers? Nature Genetics 47:8-12 (2014).) ↩
- (Li Y. et al., CRISPR Reveals a Distal Super-Enhancer Required for Sox2 Expression in Mouse Embryonic Stem Cells. PLoS One 9:e114485 (2014).) ↩
- (Hay D. et al., Genetic dissection of the α-globin super-enhancer in vivo. Nature Genetics 48:895–903 (2016).) ↩
Comments !