Enhancer Journal Club: Enhancer runaway and the evolution of diploid gene expression
I came across this very interesting theoretical paper about enhancer evolution a couple of months ago1 and thought it was worth a little discussion. The paper comes from Frederic Fyon and others at the CNRS in Montpellier. The original paper can be found at PLOS Genetics, and it's well worth a read.
Here's the abstract:
Evidence is mounting that the evolution of gene expression plays a major role in adaptation and speciation. Understanding the evolution of gene regulatory regions is indeed an essential step in linking genotypes and phenotypes and in understanding the molecular mechanisms underlying evolutionary change. The common view is that expression traits (protein folding, expression timing, tissue localization and concentration) are under natural selection at the individual level. Here, we use a theoretical approach to show that, in addition, in dip- loid organisms, enhancer strength (i.e., the ability of enhancers to activate transcription) may increase in a runaway process due to competition for expression between homologous enhancer alleles. These alleles may be viewed as self-promoting genetic elements, as they spread without conferring a benefit at the individual level. They gain a selective advantage by getting associated to better genetic backgrounds: deleterious mutations are more efficiently purged when linked to stronger enhancers. This process, which has been entirely overlooked so far, may help understand the observed overrepresentation of cis-acting regu- latory changes in between-species phenotypic differences, and sheds a new light on investigating the contribution of gene expression evolution to adaptation.
In this paper, the authors introduce and explore the concept of "enhancer runaway". The idea here is that in diploid species, the homologous chromosomes can carry different variants of the same enhancer. Selection operates on these two different enhancer alleles and under certain circumstances this can lead to competition for gene expression between the enhancers.
In the first model the authors explore, there are two chromosomes each carrying a gene driven by a single enhancer. The authors assume that the overall expression level of the gene is tightly controlled by a feedback loop, such that deleterious mutations in either enhancer do not affect the overall expression level of the gene, and only effect the allelic imbalance (i.e. how much of the overall expression is contributed by the parental or the maternal allele). I recommend browsing through the whole paper for the full results, but the conclusion the authors draw is that, under some circumstances, selection continually favours stronger enhancers.
This is in part because stronger enhancers lead to transiently imbalanced expression, with more of the protein product being generated from the gene on the same chromosome as the stron enhancer. During this period of imbalance, deleterious mutations in the linked gene have stronger effects (because they contribute to more than 50% of the total protein level) and are therefore more efficiently purged from the population. The result is that stronger enhancers find themselves more frequently linked to favourable genetic backgrounds. This means that enhancer alleles become locked in an evolutionary arms race where they continually co-evolve to be "stronger", a process that the authors term "enhancer runaway".
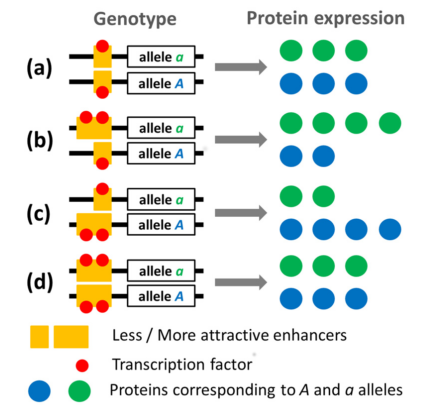
Fig 1 The strengths of two enhancer alleles are represented by their ability to attract transcription factors. Four genotypes are represented: weaker enhancer homozygote (a), stronger enhancer homozygote (d) and enhancer locus heterozygotes (b) and (c). In enhancer locus heterozygotes, the stronger enhancer is either associated with the deleterious gene allele (b) or with the viable gene allele (c). Note that in this model the total amount of proteins produced is constant. Figure reproduced with modifications from Fyon, F. et al., Enhancer Runaway and the Evolution of Diploid Gene Expression. PLOS Genetics 11 e1005665 (2015).
Of course not all genes are embedded in robust feeback loops that so stringently control their expression level. In other cases, mutations in enhancers may alter the overall expression level of a gene in addition to the imbalance between the two alleles. In this case, mutations in enhancers can decrease fitness by causing the gene to be expressed at an inappropriately high or low level. The authors consider two additional models in which gene expression level is allowed to vary and is subject to stabilizing selection (i.e. organisms that express the gene at a level higher or lower than the optimum are less fit). They find that stabilizing selection indeed slows down the process of enhancer runaway, but does not stop it completely. This is because other genes in the same pathway can co-evolve to maintain the correct gene dosage, or regulatory TFs can co-evolve to maintain the correct absolute expression levels.
The aspect of the paper I found most interesting was the exploration of recombination rate. The authors show that enhancer runaway is stronger the more close the gene and enhancer lie on the genome, because the closer they lie the less frequently they undergo recombination. This linear distance effect is greater the stronger the selection pressure on the gene. The authors use this to make a prediction that genes undergoing the strongest selection should exhibit larger regulatory regions. This reminded me of some previous work examining the regulatory regions around master developmental transcription factors, which tend to be very large and devoid of other genes2.
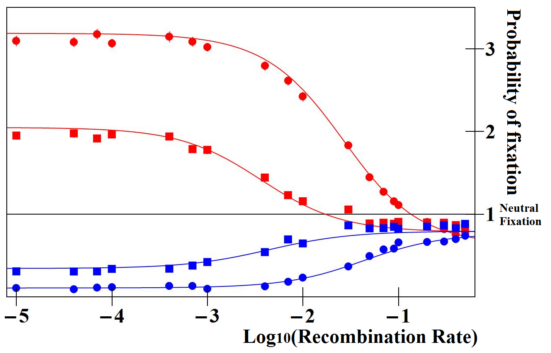
Fig 3 Ratio of fixation probabilities of mutations altering enhancer strength relative to that of a neutral mutation. In red, the mutant enhancer is three times stronger than the wild type; and in blue, three times weaker. Values are reported for the case where enhancer strength evolution does not alter overall protein expression, for various recombination rates between the enhancer and the gene (x-axis), for weak (squares) or strong selection (circles). Weaker enhancers (blue) are always selected against, while stronger enhancers (red) are selectively favored provided that they are closely linked to the gene, and disfavored otherwise. Figure reproduced from Fyon, F. et al., Enhancer Runaway and the Evolution of Diploid Gene Expression. PLOS Genetics 11 e1005665 (2015).
The authors models allow them to explore the effect of different recombination rates on the enhancer runaway process. I think it would be very interesting to explore versions of these models where the recombination rate itself was under selective pressure. Since the authors show that enhancer runaway can actually decrease fitness for the organism, perhaps they could identify conditions in which increased distance between genes and enhancer would be favoured by evolution. In other words, perhaps part of the reason why many enhancers in mammalian genomes lie at fairly large distances from their target genes3 is to reduce these enhancer runaway effects.
- (Fyon, F. et al., Enhancer Runaway and the Evolution of Diploid Gene Expression. PLOS Genetics 11 e1005665 (2015).) ↩
- (Akalin A et al. Transcriptional features of genomic regulatory blocks. Genome Biol. 10: R38 (2009)) ↩
- (Lettice, L. A. et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. PNAS. 99(11) 7548-7553.) ↩
Comments !